Focus on MPG publications¶
Imports¶
import pandas as pd
import os
import regex
import re
import string
import difflib
import numpy as np
from tqdm import tqdm
from functools import partial
from multiprocessing import Pool, cpu_count
from matplotlib.backends.backend_pdf import PdfPages
import matplotlib.pyplot as plt
Setup paths¶
gitDataPath = '../data/processedData/'
dataPath = '/media/arbeit/b88b17b8-b5d7-4c4e-86bc-ff4b67458842/sciGraph/'
General stopword routine to re-use at every step¶
rePunct = "\(|\)|\.|\d+|,|-"
rePunct
'\\(|\\)|\\.|\\d+|,|-'
stopwords = [
'Forschungsstelle',
'Förderung',
'förderung',
'Wissenschaften',
'Kaiser-Wilhelm-Institut',
'Max-Planck-gesellschaft',
'Max-Planck-Institut',
'maxplanckgesellschaft',
'maxplanckinstitut',
'maxplanckinstituts',
'maxplanckinstitute',
'maxplanckinsitut',
'maxplanckinst',
'maxplanckinstitutes',
'maxplanckinstituten',
'maxplanckinsituts',
'maxplanckinstitutt',
'maxplanckinstiut',
'maxplanckinsütut',
'planckinstitut',
'insitute',
'insitut',
'maxplanckarbeitsgruppe',
'maxplanckforschungsstelle',
'Max Planck Institute',
'Max-Planck-Institute',
'Max-Planck-Institutes',
'Max-Planck-Instituts',
'Max',
'Planck',
'MPI',
'MPG',
'Instituts',
'Institutes',
'Institute',
'Institut',
'Planck-Institut',
'zur',
'ab',
'die',
'und',
'der',
'of',
'für',
'fuer',
'fur',
'f¨r',
'fr',
'für',
'for',
'\n',
'des',
'the',
'GmbH',
'in',
'von'
]
lowerStopwords = [x.lower() for x in stopwords]
def cleanStopWords(instring):
ret = []
braket = [x.split(')(') for x in instring.split(' ')]
for x in [x for y in braket for x in y]:
x = re.sub(rePunct,'',x)
x = x.strip('\n').lower()
if x not in lowerStopwords:
if x.strip() not in ret:
ret.append(x.strip())
return ' '.join([x for x in ret if x])
Loading alternative spelling of MPIs¶
Based on OCR error files by Dirk Wintergrün.
with open(gitDataPath + 'all_labels.tsv','r', encoding='utf8') as file:
lines = [x.split('\t') for x in file.readlines()]
labels = {}
for l0 in lines[1:]:
if not 'KWI' in l0[0]:
for l1 in l0[1:]:
l1 = cleanStopWords(l1)
if l1:
labels[l1] = l0[0]
Read data from files¶
The corpus is created by appyling a regex routine on the corpus file for every year. See previous Notebook 5.
dfLoad = pd.read_csv(gitDataPath + 'mpgpubs.csv', sep='\t')
dfLoad.head(2)
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 1946 | sg:pub.10.1007/bf00643799 | Kondensationsvorgänge aus überhitztem Arsenikd... | ['de'] | sg:journal.1018189 | The Science of Nature | False | sg:person.013544521043.99;Korb_A. | https://www.grid.ac/institutes/grid.418028.7;h... |
| 1 | 1946 | sg:pub.10.1007/bf00624523 | Uber die Phosphorylase der Leukocyten | ['en'] | sg:journal.1018189 | The Science of Nature | False | Rohdewald_Margarete | https://www.grid.ac/institutes/grid.418441.c |
dfMPGall = dfLoad.drop('6',axis=1).rename(columns={'0':'year','1':'pubID','2':'title','3':'lang','4':'journalID','5':'journalName','7':'authors','8':'affName'})
dfMPGall.head(2)
| year | pubID | title | lang | journalID | journalName | authors | affName | |
|---|---|---|---|---|---|---|---|---|
| 0 | 1946 | sg:pub.10.1007/bf00643799 | Kondensationsvorgänge aus überhitztem Arsenikd... | ['de'] | sg:journal.1018189 | The Science of Nature | sg:person.013544521043.99;Korb_A. | https://www.grid.ac/institutes/grid.418028.7;h... |
| 1 | 1946 | sg:pub.10.1007/bf00624523 | Uber die Phosphorylase der Leukocyten | ['en'] | sg:journal.1018189 | The Science of Nature | Rohdewald_Margarete | https://www.grid.ac/institutes/grid.418441.c |
Resources for Sektion mapping¶
Person DB¶
One version was manually cleaned to delete double mentions, years, brackets and so on in instituts names. This data was then used to create a dictionary of mappings.
dfInstCleaned = pd.read_csv(gitDataPath + 'mpg_institute_cleanded.csv')
preprocessed = dfInstCleaned['institut_name']
The main information about the institutes of the MPG is derived from the research database of the research program “History of the Max Planck Society”, for more details see the website.
dfInstDirty = pd.read_csv(gitDataPath + 'Institute.csv')
dfInstDirtMPG = dfInstDirty[dfInstDirty.is_mpg == True].reset_index(drop=True)
dfInstDirtMPG.insert(0,'preprocessed', preprocessed)
cleanstopDirt = dfInstDirtMPG.preprocessed.apply(lambda row: cleanStopWords(row))
dfInstDirtMPG.insert(0,'cleaned',cleanstopDirt)
Two translation dictionaries : from the cleaned words to the database institut name, and the same to the sektion. At least one MPI ( Empirical Aestetics ) has no Sektion.
input2DBName = dfInstDirtMPG[['cleaned','institut_name','date_founded']].sort_values('date_founded').set_index('cleaned')['institut_name'].to_dict()
input2Sektion = dfInstDirtMPG[['cleaned','sektion','date_founded']].sort_values('date_founded').replace('','MPG').fillna('None').set_index('cleaned')['sektion'].to_dict()
Additionaly, create dictionary to translate from database name to sektion.
dbname2Sektion = dfInstDirtMPG[['institut_name','sektion','date_founded']].sort_values('date_founded').replace('','MPG').fillna('None').set_index('institut_name')['sektion'].to_dict()
Manual translations of OCR errors¶
This list is created manually, by comparing likely hits of instituts names.
dfManualTransl = pd.read_csv(gitDataPath + 'translations_inst_names.csv')
dfManualTransl.insert(0,'cleaned',dfManualTransl['0'].apply(lambda row: cleanStopWords(row)))
ocrInput2DBName = dfManualTransl[['cleaned','Unnamed: 0']].drop_duplicates().set_index('cleaned')['Unnamed: 0'].to_dict()
def findKey(inputString,debug=False,debugStr=''):
ret = ''
l1 = cleanStopWords(inputString)
if l1 in input2DBName.keys():
ret = input2DBName[l1]
elif l1 in labels.keys():
l2 = cleanStopWords(labels[l1])
if l2 in input2DBName.keys():
ret = input2DBName[l2]
else:
ret=inputString
if debug:
return f'{debugStr}: Input: {inputString} Output: {ret}'
else:
return ret
Grid data¶
Source : Global Research Identifier Database
This data is used to map between identifiers of the form grid.45667.2 to instituts name. All MPG instituts are related to the MPG (GridID grid.4372.2). This allows also to create lists of german to english translations of names.
dfGrid = pd.read_csv(dataPath + 'grid/grid-2020-06-29/grid.csv')
dfGridRelations = pd.read_csv(dataPath + 'grid/grid-2020-06-29/full_tables/relationships.csv')
dfGRIDLabels = pd.read_csv(dataPath + 'grid/grid-2020-06-29/full_tables/labels.csv')
mpgGridIDs = dfGridRelations[dfGridRelations.grid_id == 'grid.4372.2'].related_grid_id.values
name2GRID = dfGRIDLabels[dfGRIDLabels.grid_id.isin(mpgGridIDs)][['grid_id','label']].set_index('label')['grid_id'].to_dict()
grid2name = {y:x for x, y in name2GRID.items()}
If there are no german labels, use the main data.
for grid_i in mpgGridIDs:
if grid_i not in grid2name.keys():
grid2name.update({grid_i:dfGrid[dfGrid.ID == grid_i].Name.values[0]})
English translations to DB Names¶
dfGridEngl = dfGrid[dfGrid.ID.isin(mpgGridIDs)][['ID','Name']]
germ = dfGridEngl.ID.apply(lambda row: grid2name[row])
dfGridEngl.insert(2,'gerName',germ)
cleanEng = dfGridEngl.Name.apply(lambda row: cleanStopWords(row))
dfGridEngl.insert(2,'cleanEng',cleanEng)
transl = dfGridEngl.gerName.apply(lambda row: findKey(row,debug=False))
dfGridEngl.insert(3,'foundMPIs',transl)
enginput2DBName = dfGridEngl[['cleanEng','foundMPIs']].set_index('cleanEng')['foundMPIs'].to_dict()
Finding the right MPIs¶
This routine uses regular expression to find the affiliated MPI. In the SciGraph data are three main cases of affiliation data.
If the affiliation can be disambiguated, the GRID ID is used. If all affiliations can be disambiguated, you find expressions
https://grid.ac/institutes/grid.419696.5joined by semikolon. For each grid id, we check if it is part of the MPG list.For older publications, often no disambiguation was possible, such that the affiliation string has the form
"'affil 1','affil 2',' affil 3'". Here, regular expressions are used to find names of MPIs. Since the exact form of the string varies a lot, we have a long list of possible spellings and also OCR errors.For mixed cases of disambiguated and raw affiliation data, we check for each element, if grid or not and then apply the relevant method.
In each case, the found MPI is converted to terms with the cleaning routine above. Then various dictionaries are aplied to find the correct database institut. Results are joined by a semikolon.
Known problems:
For some MPIs the affiliation could be captured by
([^'\[])+Max\sPlanck\sInstitute,([^;,]+)+, i.e. the name of the institute comes before mentioning that its a MPI. These cases lead to missing all other cases, such that they have to be captures manually after the main routine.Some special collaborations, like the
MPG-CASpartner instituts are not captured.Some miss detection exists, like
MPG Ranch.
findMPI = """
Max-Delbrück-Laboratorium\sin\sder\sMPG|
Klinische\sArbeitsgruppe\sder\sMPG|
MPG\sArbeitsgruppe\s([^;,]+)+| #
MPG-AG\s([^;,]+)+| #
MPG-CAS\sPICB|#
CAS-MPG\sPartner\sInstitute|#
Fritz-Haber\sInstitut\sder\sMPG| #
Fritz-Haber-Institut\sof\sthe\sMPG| #
Fritz-Haber-Institute\sder\sMPG|#
Fritz-Haber-Institut\s{1,2}der\sMPG|#
Deutschen\sForschungs-anstalt\sfür\sPsychiatrie| #
Deutschen\sForschungsanstalt\sfür\sPsychiatrie| #
Deutsche\sForschungsanstalt\sfür\sPsychiatrie| #
KWI\sfür([^;,]+)+| #
MPI\sfür([^;,]+)+| #
MPI\sfor([^;,]+)+| #
MPI\szur([^;,]+)+|#
MPI\sof([^;,]+)+|#
MPI\s([^;,]+)+|#
Max-Planck-Institut\sf\.([^;,]+)+|
Max\sPlanck\sInstitut\sf\.([^;,]+)+|
Max-Planck-Institute\sf\.([^;,]+)+|
Max-Planck-Inst\sf\.([^;,]+)+|
Max\sPlanck\sInstitute\sf\.([^;,]+)+|
Max-Planck-Insütut\sfür([^;,]+)+|
Max\sPlanck\sInsütut\sfür([^;,]+)+|
Max-Planck-Institut\sfür([^;,]+)+|
Max-Planck-Instituts\sfür([^;,]+)+|
Max-Planck-Institute\sfür([^;,]+)+|
Max\sPlanck\sInstituts\sfür([^;,]+)+|
Max-Planck-Institutes\sfür([^;,]+)+|
Max\sPlanck\sInstitutes\sfür([^;,]+)+|
Max\sPlanck\sInstitut\sfür([^;,]+)+|#
Max\sPlanck\sInstitute\sfür([^;,]+)+|#
Max-Planck-Instituten\sfür([^;,]+)+|#
Max\sPlanck\sInstituten\sfür([^;,]+)+|#
Max\sPlanck\sInstitute\sfuer([^;,]+)+|#
Max\sPlanck\sInstitut\sfuer([^;,]+)+|#
Max-Planck-Institut\sfuer([^;,]+)+|#
Max-Planck-Institute\sfuer([^;,]+)+|#
Max-Planck-Institut\sf\?r([^;,]+)+|#
Max\sPlanck\sInstitut\sf\?r([^;,]+)+|#
Max-Planck-Institut\szur([^;,]+)+|#
Max\sPlanck\sInstitut\szur([^;,]+)+|#
Max\sPlanck\sInstitute\sfor([^;,]+)+| #
Max-Planck-Institute\sfor([^;,]+)+| #
Max-Planck-Institut\sfor([^;,]+)+| #
Max\sPlanck\sInstitute\sof([^;,]+)+| #
Max-Planck-Institute\sof([^;,]+)+| #
Max-Planck-Institut\sof([^;,]+)+| #
Max-Planck-Institute\s([^;,]+)+| #
Max\sPlanck\sInstitute\s([^;,]+)+| #
Max-Planck-Insitut\sfür([^;,]+)+| #
Max\sPlanck\sInsitut\sfür([^;,]+)+| #
Max-Planck-Institut\sfur([^;,]+)+| #
Max\sPlanck\sInstitut\sfur([^;,]+)+| #
Max-Planck-Inst.\sfür([^;,]+)+| #
Max\sPlanck\sInst\.\sfür([^;,]+)+| #
Max-Planck-Inst\.\sf\.([^;,]+)+| #
Max\sPlanck\sInst\.\sf\.([^;,]+)+| #
Max-Planck-Instiut\sfür([^;,]+)+| #
Max\sPlanck\sInstiut\sfür([^;,]+)+| #
Max-Planck-Insituts\sfür([^;,]+)+| #
Max\sPlanck\sInsituts\sfür([^;,]+)+| #
Max-Planck-Institut\s([^;,]+)+| #
Max\sPlanck\sInstitut\s([^;,]+)+| #
Max\sPlanck\sInsitute\sfor\s([^;,]+)+| #
Max-Planck-Institutt\sfür\s([^;,]+)+#
"""
find = re.compile(findMPI,re.X|re.MULTILINE)
def findKeyString(inputString,debug=False,debugStr=''):
ret = ''
l1 = cleanStopWords(inputString)
if l1 in input2DBName.keys():
ret = input2DBName[l1]
elif l1 in enginput2DBName.keys():
ret = enginput2DBName[l1]
elif l1 in ocrInput2DBName.keys():
ret = ocrInput2DBName[l1]
elif l1 in labels.keys():
l2 = cleanStopWords(labels[l1])
if l2 in input2DBName.keys():
ret = input2DBName[l2]
elif l2 in enginput2DBName.keys():
ret = enginput2DBName[l2]
elif l2 in ocrInput2DBName.keys():
ret = ocrInput2DBName[l2]
else:
ret=inputString
if debug:
return f'{debugStr}: Input: {inputString} Output: {ret}'
else:
return ret
def cleanInput(text, debug = False):
ret = []
parts = text.split(';')
if len(parts) == 1:
splitted = regex.sub("'","#", text.strip(']|[')).split('#, #')
for part in splitted:
exp1 = regex.sub('#|\*|\(|\)|\[|\]|\+|\{|\}|\n', '', part)
res = re.search(find, exp1)
if res:
try:
ret.append(findKeyString(res.group(0), debug=debug, debugStr='regexNoGrid'))
except:
raise
else:
for part in parts:
if part.startswith('http'):
try:
gridid = part.split("https://www.grid.ac/institutes/")[1].strip('\n')
ret.append(findKeyString(grid2name[gridid], debug=debug, debugStr='grid'))
except KeyError:
pass
except IndexError:
pass
except:
raise
else:
exp1 = regex.sub('#|\*|\(|\)|\[|\]|\+|\{|\}|\n', '', part)
res = re.search(find, exp1)
if res:
try:
ret.append(findKeyString(res.group(0), debug=debug, debugStr='regexGrid'))
except:
raise
return ';'.join(ret)
out = dfMPGall.affName.apply(lambda row: cleanInput(row))
for col in ['foundMPIs','sektion']:
if col in dfMPGall.columns:
dfMPGall = dfMPGall.drop(col,axis=1)
dfMPGall.insert(0,'foundMPIs',out)
Find corresponding Sektion¶
Using the found database institute names, we can now also find the Sektion.
error = []
possibleVals = []
def applyDict(row,dbname=dbname2Sektion,di=input2Sektion, debug=False):
res = ['None' for i,j in enumerate(row.split(';'))]
for idx, part in enumerate(row.split(';')):
if part in dbname.keys():
if debug:
res[idx] = f'dbname2sek: {dbname[part]}'
else:
res[idx] = dbname[part]
else:
cl = cleanStopWords(part)
if cl in di.keys():
if debug:
res[idx] = f'input2sek_lev1: {di[cl]}'
else:
res[idx] = di[cl]
elif cl in labels.keys():
try:
l2 = cleanStopWords(labels[cl])
if debug:
res[idx] = f'input2sek_lev2:cl:{cl} l2:{l2} res:{di[l2]}'
else:
res[idx] = di[l2]
except:
raise
else:
if debug:
resMPIs = []
for pcl in cl.split():
for x in input2DBName.keys():
if pcl in x.split():
resMPIs.append(input2DBName[x])
for x in enginput2DBName.keys():
if pcl in x.split():
resMPIs.append(enginput2DBName[x])
for x in ocrInput2DBName.keys():
if pcl in x.split():
resMPIs.append(ocrInput2DBName[x])
possibleVals.append((part,cl,resMPIs))
error.append((cl))
try:
return ';'.join(res)
except:
print(res)
sektion = dfMPGall.foundMPIs.apply(lambda row: applyDict(row, debug=False))
dfMPGall.insert(1,'sektion',sektion)
How many author contributions can be found for each Sektion.
sektion.apply(lambda row: row.split(';')).explode().value_counts()
CPTS 71771
BMS 51065
None 8292
GSHS 5715
MPG 10
Name: foundMPIs, dtype: int64
dfMPGall.shape
(53446, 10)
Distribution of author numbers in the Sektionen¶
authBMS = dfMPGall[dfMPGall.sektion.str.contains('BMS')].authors.fillna('').apply(lambda row: len(row.split(';'))).to_frame().reset_index(drop=True).value_counts().to_frame().rename(columns={0:'BMS'})
authCPTS = dfMPGall[dfMPGall.sektion.str.contains('CPTS')].authors.fillna('').apply(lambda row: len(row.split(';'))).to_frame().reset_index(drop=True).value_counts().to_frame().rename(columns={0:'CPTS'})
authGSHS = dfMPGall[dfMPGall.sektion.str.contains('GSHS')].authors.fillna('').apply(lambda row: len(row.split(';'))).to_frame().reset_index(drop=True).value_counts().to_frame().rename(columns={0:'GSHS'})
dfCoauthorenSektionen = authBMS.merge(authCPTS,left_index=True, right_index=True,how='outer').merge(authGSHS,left_index=True, right_index=True,how='outer')
dfCoauthorenSektionen
| BMS | CPTS | GSHS | |
|---|---|---|---|
| authors | |||
| 1 | 1895.0 | 523.0 | 86.0 |
| 2 | 4730.0 | 4660.0 | 697.0 |
| 3 | 3777.0 | 4101.0 | 558.0 |
| 4 | 2685.0 | 2906.0 | 378.0 |
| 5 | 1947.0 | 1874.0 | 265.0 |
| ... | ... | ... | ... |
| 3173 | NaN | 1.0 | NaN |
| 3180 | NaN | 1.0 | NaN |
| 3195 | NaN | 1.0 | NaN |
| 5100 | NaN | 2.0 | NaN |
| 5114 | NaN | 2.0 | NaN |
538 rows × 3 columns
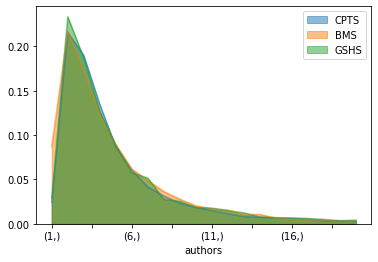
bmsNorm = dfCoauthorenSektionen['BMS']/dfCoauthorenSektionen.BMS.sum()
cptsNorm = dfCoauthorenSektionen['CPTS']/dfCoauthorenSektionen.CPTS.sum()
gshsNorm = dfCoauthorenSektionen['GSHS']/dfCoauthorenSektionen.GSHS.sum()
pd.DataFrame([dfCoauthorenSektionen['CPTS'],dfCoauthorenSektionen['BMS'],dfCoauthorenSektionen['GSHS']]).transpose()[:20].plot.area(stacked=False)
/home/arbeit/Dokumente/gwdgGitlab/GMPG/gmpg-notebooks/env/lib/python3.8/site-packages/pandas/plotting/_matplotlib/core.py:1235: UserWarning: FixedFormatter should only be used together with FixedLocator
ax.set_xticklabels(xticklabels)
<AxesSubplot:xlabel='authors'>
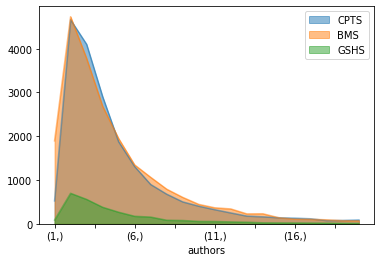
pd.DataFrame([cptsNorm,bmsNorm,gshsNorm]).transpose()[:20].plot.area(stacked=False)
/home/arbeit/Dokumente/gwdgGitlab/GMPG/gmpg-notebooks/env/lib/python3.8/site-packages/pandas/plotting/_matplotlib/core.py:1235: UserWarning: FixedFormatter should only be used together with FixedLocator
ax.set_xticklabels(xticklabels)
<AxesSubplot:xlabel='authors'>
Publications without Sektion data¶
For some publications, the contribution of an MPI is clear, but finding the right institute requieres expert knowledge. This work can be done in the cleaning app, developed by SHKs.
dfNotFound = dfMPGall[dfMPGall.sektion == 'None']
for val in dfNotFound.foundMPIs.unique():
print(val)
Max-Planck-Institut München
Max-Planck-Institut in München
Max Planck Institut in Berlin
MPI Garching
Max-Planck-Institut HML
Max Planck Institut Göttingen
Deutsches Klimarechenzentrum
Forschungszentrum caesar
MPI für Verhaltensbiologie
MPI Bonn
MPI Mines Ltd.
Max Planck Institute Jena
Gesellschaft für wissenschaftliche Datenverarbeitung mbH Göttingen
MPI Research Inc.
MPI - CNRS UMR5146 - Centre SMS
Max Planck Digital Library
Max-Planck-Institut für Herz- und Lungenforschung
Center for Free-Electron Laser Science
MPI for Bioinformatics
MPI Inc.
Beilstein-Institut zur Förderung der Chemischen Wissenschaften
CAS-MPG Partner Institute
MPI Academy
Max Planck Graduate Center
Max Planck Florida Institute for Neuroscience
Ernst Strüngmann Institut
Max-Planck-Institut für Immunobiologie und Epigenetik
Max Planck Innovation
MPI für empirische Ästhetik
Max Planck Institute Luxembourg
MPI Leipzig
MPI Saarbrücken
Max Planck Institute Luxemburg for International, European and Regulatory Procedural Law
MPI ANR-11-LABX-0007-01
MPI Tübingen
Export to file¶
To create a new exported version, uncomment the following line, and change the file extension to the current date.
dfMPGall.head(2)
| foundMPIs | sektion | year | pubID | title | lang | journalID | journalName | authors | affName | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Fritz-Haber-Institut der MPG;Fritz-Haber-Insti... | CPTS;CPTS | 1946 | sg:pub.10.1007/bf00643799 | Kondensationsvorgänge aus überhitztem Arsenikd... | ['de'] | sg:journal.1018189 | The Science of Nature | sg:person.013544521043.99;Korb_A. | https://www.grid.ac/institutes/grid.418028.7;h... |
| 1 | None | 1946 | sg:pub.10.1007/bf00624523 | Uber die Phosphorylase der Leukocyten | ['en'] | sg:journal.1018189 | The Science of Nature | Rohdewald_Margarete | https://www.grid.ac/institutes/grid.418441.c |
#dfMPGall.to_csv(gitDataPath + 'MPGPubSektionen_11112020.tsv',sep='\t',index=False)
Some first interpretations¶
Intra-Sektional collaborations¶
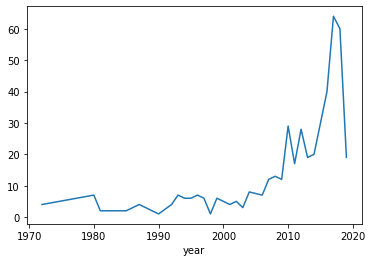
Publications with collaborations of authors from more then once Sektion are relatively rare until the early 2000thst.
dfMPGall[dfMPGall.sektion.apply(lambda row: len(set(row.split(';')))>1)].groupby('year').size().plot()
<AxesSubplot:xlabel='year'>
intraCollab = dfMPGall[dfMPGall.sektion.apply(lambda row: len(set([x for x in row.split(';') if x != 'None']))>1)]
intraCollab.foundMPIs.apply(lambda row: row.split(';')).explode().value_counts().to_frame()
| foundMPIs | |
|---|---|
| MPI für biophysikalische Chemie (Karl-Friedrich-Bonhoeffer-Institut) (seit 1971) | 259 |
| MPI für Psychiatrie (bis 1966 Deutsche Forschungsanstalt für Psychiatrie (MPI)) | 195 |
| MPI für experimentelle Medizin (1948-1965 Medizinische Forschungsanstalt der MPG) | 149 |
| MPI für medizinische Forschung | 123 |
| Center for Free-Electron Laser Science | 122 |
| ... | ... |
| MPI für Struktur und Dynamik der Materie | 1 |
| MPI für Menschheitsgeschichte (ab 2014) | 1 |
| MPI für die Physik des Lichts | 1 |
| MPI für bioanorganische Chemie (2003-2012) | 1 |
| MPI für Verhaltensphysiologie (1954-2003) | 1 |
63 rows × 1 columns
There is only one publication from all three Sektionen, published 2018.
dfMPGall[dfMPGall.sektion.apply(lambda row: len(set([x for x in row.split(';') if x != 'None']))>2)]
| foundMPIs | sektion | year | pubID | title | lang | journalID | journalName | authors | affName | |
|---|---|---|---|---|---|---|---|---|---|---|
| 49894 | MPI für biologische Kybernetik;MPI für biologi... | BMS;BMS;CPTS;BMS;GSHS;GSHS;BMS;BMS | 2018 | sg:pub.10.1038/s41467-018-06304-z | LISA improves statistical analysis for fMRI | ['en'] | sg:journal.1043282 | Nature Communications | sg:person.01110373331.54;sg:person.01323631320... | https://www.grid.ac/institutes/grid.419501.8;h... |
Output per Sektion¶
dfMPGall.columns
Index(['foundMPIs', 'sektion', 'year', 'pubID', 'title', 'lang', 'journalID',
'journalName', 'authors', 'affName'],
dtype='object')
sekSp = dfMPGall.sektion.apply(lambda row: row.split(',')[0].split(';'))
dfMPGall.insert(0,'sekSplit', sekSp)
dfMPGall.sekSplit.iloc[3]
['CPTS', 'CPTS']
result = []
for year, g0 in dfMPGall.groupby('year'):
res = {'year':year}
for sektion, g1 in g0.explode('sekSplit').groupby('sekSplit'):
res[sektion] = g1.shape[0]
result.append(res)
#result
dfSektionOutput = pd.DataFrame(result)[:-15]
dfSektionOutput = dfSektionOutput.set_index('year')
dfSektionOutput = dfSektionOutput.rename(columns={'None':'Keine Sektion'})
dfSektionOutput.columns = sorted(dfSektionOutput.columns)
dfSektionOutput.index.rename('Jahr',inplace=True)
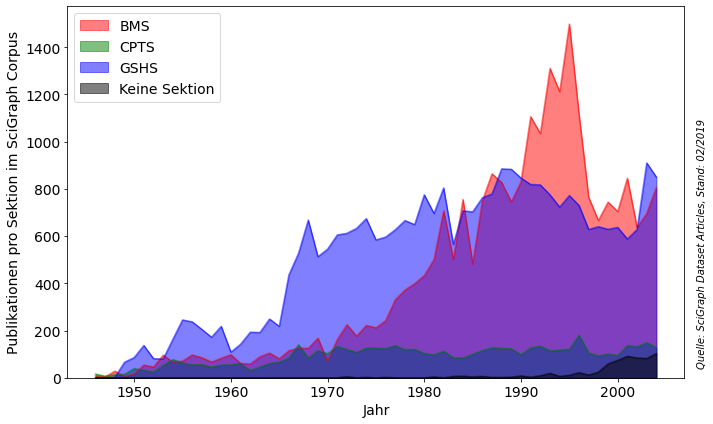
colorSektions = ['r','g','b','k']
with PdfPages('../results/Fig8_MPG_Sektionen.pdf') as pdffigure:
fig, ax1 = plt.subplots(figsize=(10,6))
color = 'k'
ax1.set_xlabel('Jahr',fontsize = 14)
ax1.set_ylabel('Publikationen pro Sektion im SciGraph Corpus', color=color,fontsize = 14)
plt1 = dfSektionOutput[['BMS','CPTS','GSHS','Keine Sektion']].plot.area(stacked=False, ax=ax1, color=colorSektions)
ax1.tick_params(axis='y', labelcolor=color, labelsize = 14)
ax1.tick_params(axis='x', labelcolor=color, labelsize = 14)
ax1.legend(fontsize = 14,loc=2)
ax1.annotate(
"Quelle: SciGraph Dataset Articles, Stand: 02/2019", (
1.02,0.7),(0, 0), xycoords = "axes fraction", textcoords = "offset points", va = "top", style = "italic", fontsize = 10, rotation = 90)
fig.tight_layout()
# Save as PDF
#pdffigure.savefig(bbox_inches="tight", dpi=400)
# Save as PNG
#plt.savefig('../../../results/SciGraph/SciGraph_MPG_Publikationen_Sektionen.png')
plt.show()
plt.close()