Analysis
Contents
Analysis¶
Build base corpus for semanticlayertools¶
import os
import requests
from semanticlayertools.linkage.cocitation import Cocitations
from semanticlayertools.clustering.leiden import TimeCluster
from semanticlayertools.clustering.reports import ClusterReports
from semanticlayertools.visual.utils import streamgraph
import pandas as pd
from tqdm import tqdm
import dimcli
dimcli.login()
dsl = dimcli.Dsl()
Searching config file credentials for default 'live' instance..
Dimcli - Dimensions API Client (v0.9.6)
Connected to: <https://app.dimensions.ai/api/dsl> - DSL v2.0
Method: dsl.ini file
This routine makes use of the Research Organization Registry which contains data for relation among research organizations. This allows to query for sub-organizations in relation to a parent. The baseOrgID in this case is the Max Planck Society: https://ror.org/01hhn8329
orgName = "MPG"
baseOrgID = "01hhn8329"
In this notebook we are interested in an earlier phase of the MPG and therefore limit the query for publications.
yearStart = 1945
yearEnd = 1970
yearRange = range(yearStart, yearEnd, 1)
basepath = "./basedata/"
Determine grid IDs of children for parent organization¶
Since DimensionsAI makes use of the GRID identifiers, we first create a list of GRID IDs for all children organizations of the MPG.
baseurl = "https://api.ror.org/organizations/"
res = requests.get(f'{baseurl}{baseOrgID}')
orgChildren = [x['id'] for x in res.json()['relationships']]
orgChildIDs = []
for child in orgChildren:
res = requests.get(f'{baseurl}{child}')
orgChildIDs.append(
res.json()['external_ids']['GRID']['all']
)
Note that some organizations might have several IDs. This is not the case for this query however.
len(orgChildIDs)
107
Query for available publications¶
Using the DimensionsAI CLI, dimcli, we can create a query for each grid ID. If for a grid ID data is found in the given timeframe, a record is created for later processing which year, id, references, title and authors of each publication found.
%%time
for gridid in tqdm(orgChildIDs):
query = f"""search publications
where year in [{yearStart}:{yearEnd}] and research_orgs="{gridid}"
return publications[year+id+reference_ids+title+authors] sort by year"""
res = dsl.query_iterative(query, verbose=False)
dfRes = res.as_dataframe()
if dfRes.shape[0] > 0:
dfRes.to_json(f'{basepath}{gridid}.json', orient='records', lines=True)
100%|█████████████████████████████████████████| 107/107 [04:15<00:00, 2.39s/it]
CPU times: user 4.88 s, sys: 344 ms, total: 5.23 s
Wall time: 4min 15s
Preprocess base corpus¶
The retrieved data has to be reworked slightly to fit the format of the semanticlayertools package. From the author information, affiliations and fullnames of multiple authors are joined with semincolons. Then the full corpus is split into years and exported to the folder corpus.
files = [x for x in os.listdir(basepath) if x.endswith('.json')]
resDF = []
for x in files:
dfT = pd.read_json(f'{basepath}{x}', lines=True)
dfT.insert(0, 'org', x[:-5])
resDF.append(dfT)
dfall = pd.concat(resDF)
dfall.shape
(6254, 6)
dfall.head(2)
| org | authors | id | reference_ids | title | year | |
|---|---|---|---|---|---|---|
| 0 | grid.419604.e | [{'affiliations': [{'city': 'Stony Brook', 'ci... | pub.1062501435 | [pub.1062498890, pub.1062498889, pub.106249893... | Potassium-Argon Ages of Lunar Rocks from Mare ... | 1970 |
| 1 | grid.419604.e | [{'affiliations': [{'city': 'Heidelberg', 'cit... | pub.1062499600 | [pub.1044841340, pub.1003581711, pub.101588479... | Fission Track Ages and Ages of Deposition of D... | 1970 |
affStr = dfall.authors.apply(lambda row: ';'.join(['+'.join([y['name'] for y in x['affiliations']]) for x in row]) if type(row)==list else 'NoAff')
authorStr = dfall.authors.apply(lambda row: ';'.join([x['first_name'] + ' ' + x['last_name'] for x in row]) if type(row)==list else 'NoAuthor')
dfall = dfall.drop('authors', axis=1)
dfall.insert(0, 'aff', affStr)
dfall.insert(0, 'authors', authorStr)
dfall.head(2)
| authors | aff | org | id | reference_ids | title | year | |
|---|---|---|---|---|---|---|---|
| 0 | O. A. Schaeffer;J. G. Funkhouser;D. D. Bogard;... | Stony Brook University;Stony Brook University;... | grid.419604.e | pub.1062501435 | [pub.1062498890, pub.1062498889, pub.106249893... | Potassium-Argon Ages of Lunar Rocks from Mare ... | 1970 |
| 1 | W. Gentner;B. P. Glass;D. Storzer;G. A. Wagner | Max Planck Institute for Nuclear Physics;Godda... | grid.419604.e | pub.1062499600 | [pub.1044841340, pub.1003581711, pub.101588479... | Fission Track Ages and Ages of Deposition of D... | 1970 |
dfall.to_csv(f'./{orgName}_full.csv', index=False)
for year, g0 in dfall.groupby('year'):
g0[['org','year','id','authors','aff','title','reference_ids']].to_json(f'./corpus/{orgName}_{year}.json', orient="records", lines=True)
Apply cocitation clustering¶
We can now apply the clustering method for the cocitation network of this corpus. As a first step we create the network of cocitation for each year slice.
Build cocitation networks¶
The cocitation network is created by linking pairs of references, if they appear in the same publication. A link is dated with the cociting papers year.
semCocite = Cocitations(
inpath = './corpus/',
outpath = './cocitation/',
columnName = "reference_ids",
)
semCocite.processFolder()
As a first measure of the data quality, we can create a plot of the amount of nodes and edges contained in the giant component.
gcEvol = pd.read_csv('./cocitation/Giant_Component_properties.csv')
gcEvol.sort_values('year').set_index('year')[['nodespercent', 'edgepercent']].plot()
<AxesSubplot:xlabel='year'>
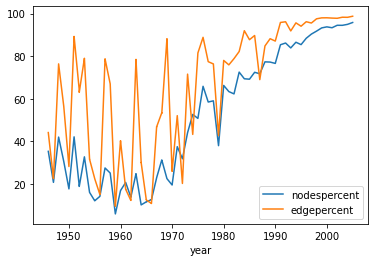
For the timeframe considered, the giant component contains only around 30 percent of all nodes. An analysis of the full data, not only the giant component, could therefore be advisable.
Find time clusters¶
timeClustering = TimeCluster(
inpath = './cocitation/',
outpath = './clusters/',
timerange = (yearStart, yearEnd)
)
Graphs between 1946 and 1970 loaded in 2.7144150733947754 seconds.
outfile, clusteredData = timeClustering.optimize()
Set layers.
Set partitions.
Set interslice partions.
Finished in 9.795211791992188 seconds.Found 76 clusters, with 1 larger then 1000 nodes.
Visualize cluster evolution¶
streamgraph(
outfile,
minClusterSize = 200,
smooth=True,
);
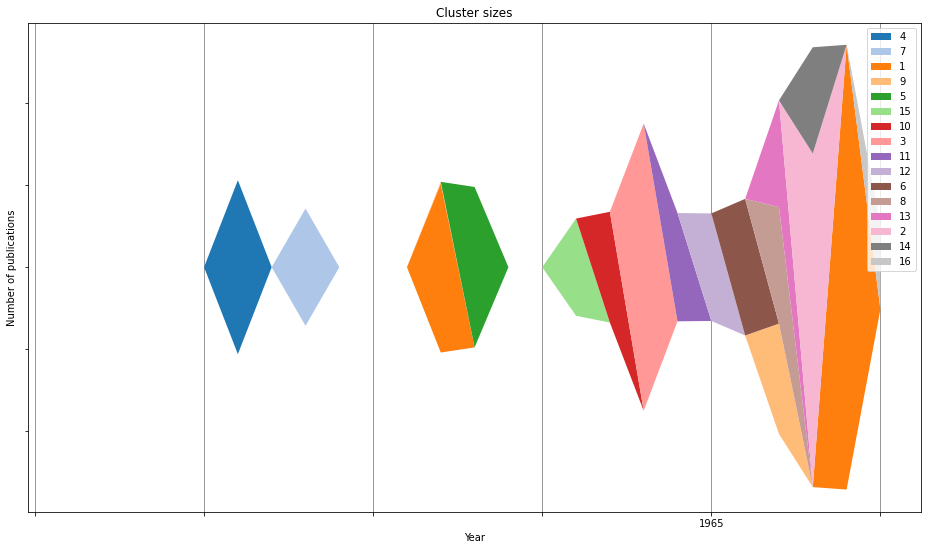
Generate cluster reports for inspection¶
generateRep = ClusterReports(
infile = outfile,
metadatapath = './corpus/',
outpath = "./reports/",
authorColumnName = "authors",
publicationIDcolumn='id',
numberProc = 4,
minClusterSize = 200,
)
generateRep.gatherClusterMetadata()
generateRep.writeReports()
! head -n 40 './reports/timeclusters_1945-1970_res_0.003_intersl_0.4/Cluster_1.txt'
Report for Cluster 1
Got 1488 unique publications in time range: 1957 to 1969.
Found metadata for 44 publications.
There are 1466 publications without metadata.
The top 20 authors of this cluster are:
Richard Kuhn: 44
Adeline Gauhe: 14
Hans Helmut Baer: 12
Irmentraut Löw: 6
Werner Kirschenlohr: 6
Heinrich Trischmann: 4
Reinhard Brossmer: 4
H. J. Haas: 2
Hans Helmut Baaer: 2
Gerd Krüger: 2
Franz Haber: 2
Heinz Tiedemann: 2
Gerd Krüuger: 2
Paul György: 2
John R.E. Hoover: 2
Catharine S. Rose: 2
Hans W. Ruelius: 2
Friedrich Zilliken: 2
Heinz Schretzmann: 2
H. Trischmann: 2
The top 20 affiliations of this cluster are:
Max Planck Institute for Medical Research: 102
Max Planck Institute for Medical Research+University of Pennsylvania+University of Pennsylvania: 14
Topics in cluster for 15 topics:
topic 0: konstitution die solanin lacto‐n‐biose konstitution der lacto‐n‐biose des der ‐trimethyl‐d‐galaktose glucose farbreaktion frauenmilch fucosido‐lactose für galaktosi‐do‐n‐acetyl‐glucosamin hydrierungen human einfluß von substituenten auf die ihre ii influenza‐virus
topic 1: ‐acetamino‐lactose synthese synthese der ‐acetamino‐lactose der human einfluß einfluß von substituenten auf die factor farbreaktion frauenmilch fucosido‐lactose für galaktosi‐do‐n‐acetyl‐glucosamin glucose hydrierungen d‐galaktal ihre ii influenza‐virus iv
topic 2: zum von abbau von tomatidin zum ‐methyl‐‐äthyl‐pyridin ‐methyl‐‐äthyl‐pyridin abbau tomatidin bifidus c glucose radioaktiver stoffwechsel umsetzung lactobacillus des die für d‐galaktal ein einfluß einfluß von substituenten auf die
